Developmental Genomics Research Group
Group Member

 Assoc. Prof. Hajime Ogino
Assoc. Prof. Hajime Ogino
Asst. Prof. Haruki Ochi
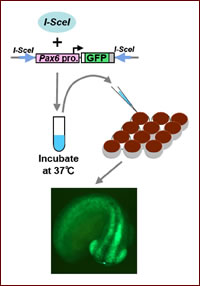
Fig. 1.High-throughput transgenesis in Xenopus tropicalis using I-SceI meganuclease. The I-SceI and transgene construct are incubated together, followed by direct injection into fertilized Xenopus eggs.
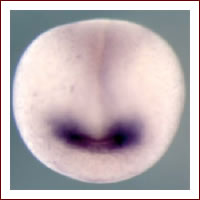
Fig. 2. Frontal view of a transgenic Xenopus gastrula embryo carrying the Six3 enhancer linked to the GFP gene. GFP expression is observed in the presumptive eye and forebrain field within the neural plate (purple staining).
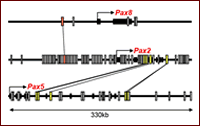
Fig. 3. Distribution of CNEs in human Pax2/5/8 loci. Red and yellow boxes indicate CNEs conserved in this paralog group.
Summary
Vertebrate genomes have a large number of conserved non-coding elements (CNEs), and initial characterization suggested that many of them are cis-regulatory elements associated with developmental regulatory genes. Since orthologous CNEs from different species often have similar but distinct cis-regulatory activities, their analysis is expected not only to reveal a conserved core of gene regulatory networks for vertebrate body plans but also to provide insights into the mechanisms that generate the morphological diversity of vertebrates. In this project, we are using a transgenic Xenopus system to investigate the regulation and roles of CNEs associated with key regulatory genes involved in head and kidney development.
Main research project
- Mechanisms of forebrain and eye development
- The presumptive forebrain and eye fields in frog embryos are formed during gastrulation by signaling from Spemann痴 organizer. We now know that BMP- and Wnt-inhibitors mediate this signaling by a mechanism that is conserved throughout vertebrates. However, how head patterning genes such as Six3 and FoxG1 are activated in response to BMP- and Wnt-inhibitors remains unknown. We are approaching this question through regulatory analyses of CNEs associated with head-patterning genes. We are also investigating relationships between species-specific enhancer activities of orthologous CNEs and species-specific morphological differences in head structures (size of telencephalon, eye, etc.).
- Evolution of cis-regulatory mechanisms following genome/gene duplication
- Vertebrate genomes have many paralogous genes that evolved from common ancestral genes by genome/gene duplication. Genes in a particular paralog group often show overlapping but distinct expression, implying that their cis-regulatory mechanisms are partially conserved. To reveal mechanisms that either generate or restrict cis-regulation diversity, we are comparing the enhancer activities of CNEs associated with paralog groups that are involved in eye, sensory placode and kidney development.
Selected publications
- Sato, S., et al., Dev. Biol., 344, 158-171, 2010
- Hellsten, U., et al., Science, 328, 633-636, 2010
- Kurokawa, D., et al., Dev. Biol., 342, 110-120, 2010
- Ochi H. & Westerfield M., BMC Dev. Biol., 9, 13, 2009
- Ogino H. & Ochi H., Dev. Growth Differ., 51, 387-401, 2009
- Sakamoto K., et al., PloS One, 4(4), e5121, 2009
- Ogino H., et al., Development, 135, 249-258, 2008
- Ochi H. et al., J. Biol. Chem., 283, 3539-3536, 2008
- Yokoyama H., et al., Dev. Biol., 306, 170-178, 2007
- Ochi H. & Westerfield M., Dev. Growth Differ., 49, 1-11, 2007
- Ogino H., et al., Nature Protocol, 1, 1703-1710, 2006
- Ogino H., et al., Mech. Dev., 123, 103-113, 2006
- Ochi H., et al., Dev. Biol., 297, 127-140, 2006
- Hirsh N., et al., Dev. Dyn., 225, 522-535, 2003
- Ochi H., et al., J. Biol. Chem., 278, 537-544, 2003
- Ogino H. & Yasuda K., Dev. Growth Differ., 42, 437-448, 2000
- Ogino H. & Yasuda K., Science, 280, 115-118, 1998
